Clinical Tables with ‘flextable’, ‘tables’ and ‘rtables’
‘flextable’ package
The primary goal of ‘flextable’(Gohel and Skintzos 2023, website) is to provide a versatile and efficient solution for creating and formatting tables in R.
It aims to make table creation and customization easier by offering a flexible and user-friendly interface within the R environment. The package offers extensive capabilities for creating table structure, format content, and appearance.
Statistic | ||
|---|---|---|
speed | Mean (SD) | 15.4 (5.3) |
Median (IQR) | 15.0 (7.0) | |
Range | 4.0 - 25.0 | |
dist | Mean (SD) | 43.0 (25.8) |
Median (IQR) | 36.0 (30.0) | |
Range | 2.0 - 120.0 |
features: quick overview
with as_flextable()
The as_flextable() function is used to transform specific objects into flextable objects.
species | island | ||||
|---|---|---|---|---|---|
Biscoe | Dream | Torgersen | Total | ||
Adelie | Count | 44 (12.8%) | 56 (16.3%) | 52 (15.1%) | 152 (44.2%) |
Mar. pct (1) | 26.2% ; 28.9% | 45.2% ; 36.8% | 100.0% ; 34.2% | ||
Chinstrap | Count | 0 (0.0%) | 68 (19.8%) | 0 (0.0%) | 68 (19.8%) |
Mar. pct | 0.0% ; 0.0% | 54.8% ; 100.0% | 0.0% ; 0.0% | ||
Gentoo | Count | 124 (36.0%) | 0 (0.0%) | 0 (0.0%) | 124 (36.0%) |
Mar. pct | 73.8% ; 100.0% | 0.0% ; 0.0% | 0.0% ; 0.0% | ||
Total | Count | 168 (48.8%) | 124 (36.0%) | 52 (15.1%) | 344 (100.0%) |
(1) Columns and rows percentages | |||||
Supported outputs
It supports various output formats to meet different needs :
- HTML
- Microsoft Word and RTF
- Microsoft PowerPoint
- ‘grid graphics’
- Excel (https://github.com/pteridin/flexlsx)
Demographic table example
adsl <- select(formatters::ex_adsl, AGE, SEX, ARM)
col_labels <- map_chr(adsl, function(x) attr(x, "label"))
summarizor(adsl, by = "ARM") |>
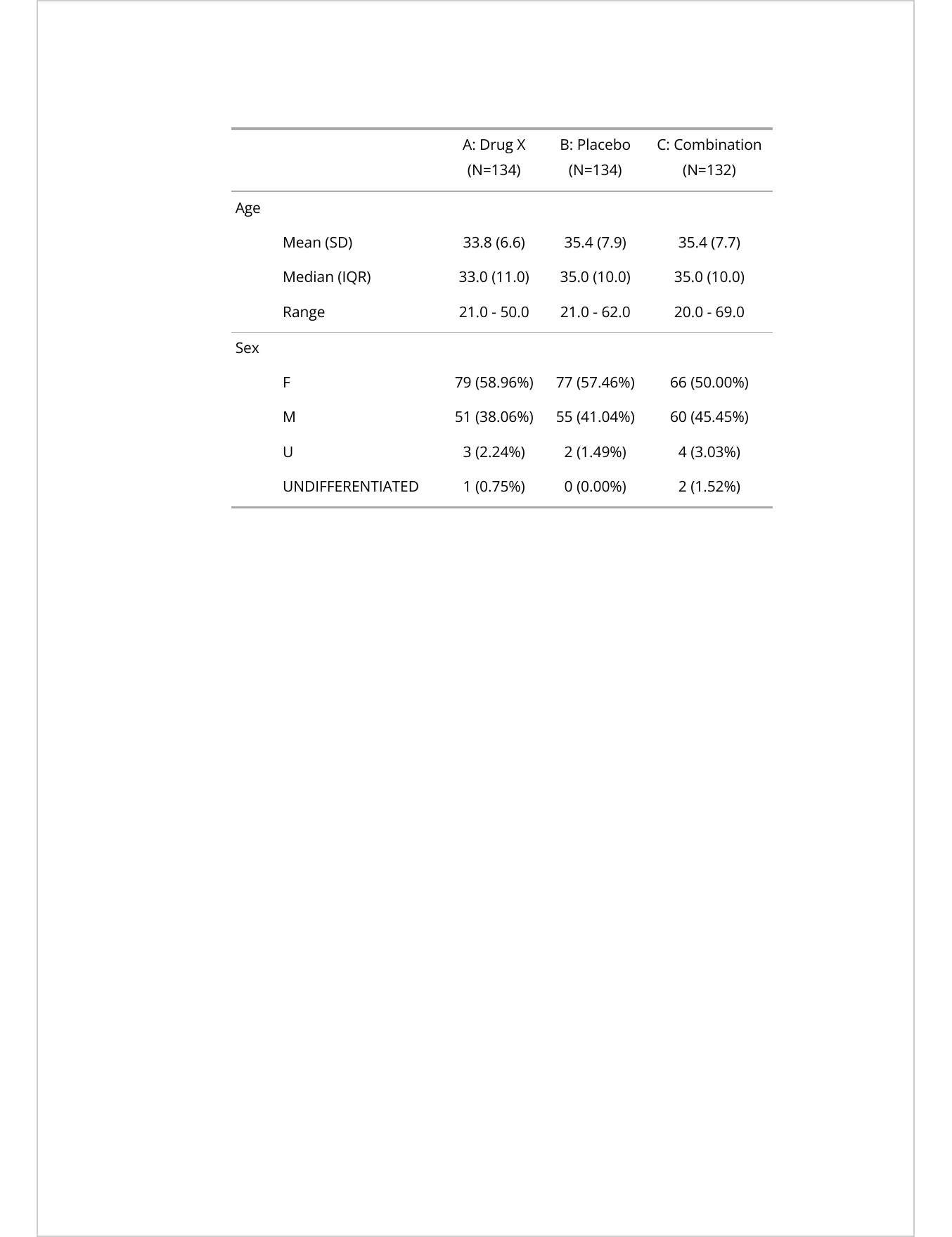
as_flextable()A: Drug X | B: Placebo | C: Combination | |||||
|---|---|---|---|---|---|---|---|
AGE | Mean (SD) | 33.8 (6.6) | 35.4 (7.9) | 35.4 (7.7) | |||
Median (IQR) | 33.0 (11.0) | 35.0 (10.0) | 35.0 (10.0) | ||||
Range | 21.0 - 50.0 | 21.0 - 62.0 | 20.0 - 69.0 | ||||
SEX | F | 79 (58.96%) | 77 (57.46%) | 66 (50.00%) | |||
M | 51 (38.06%) | 55 (41.04%) | 60 (45.45%) | ||||
U | 3 (2.24%) | 2 (1.49%) | 4 (3.03%) | ||||
UNDIFFERENTIATED | 1 (0.75%) | 0 (0.00%) | 2 (1.52%) |
Visual aspect can be improved:
- by using options of
as_flextable(), - by adding tabulations with
prepend_chunks(), - by using proper labels with
labelizor().
ft <- summarizor(adsl, by = "ARM") |>
as_flextable(sep_w = 0, separate_with = "variable",
spread_first_col = TRUE) |>
align(i = ~ !is.na(variable), align = "left") |>
prepend_chunks(i = ~ is.na(variable), j ="stat",
as_chunk("\t")) |>
labelizor(j = c("stat"), labels = col_labels,
part = "all") |>
autofit()
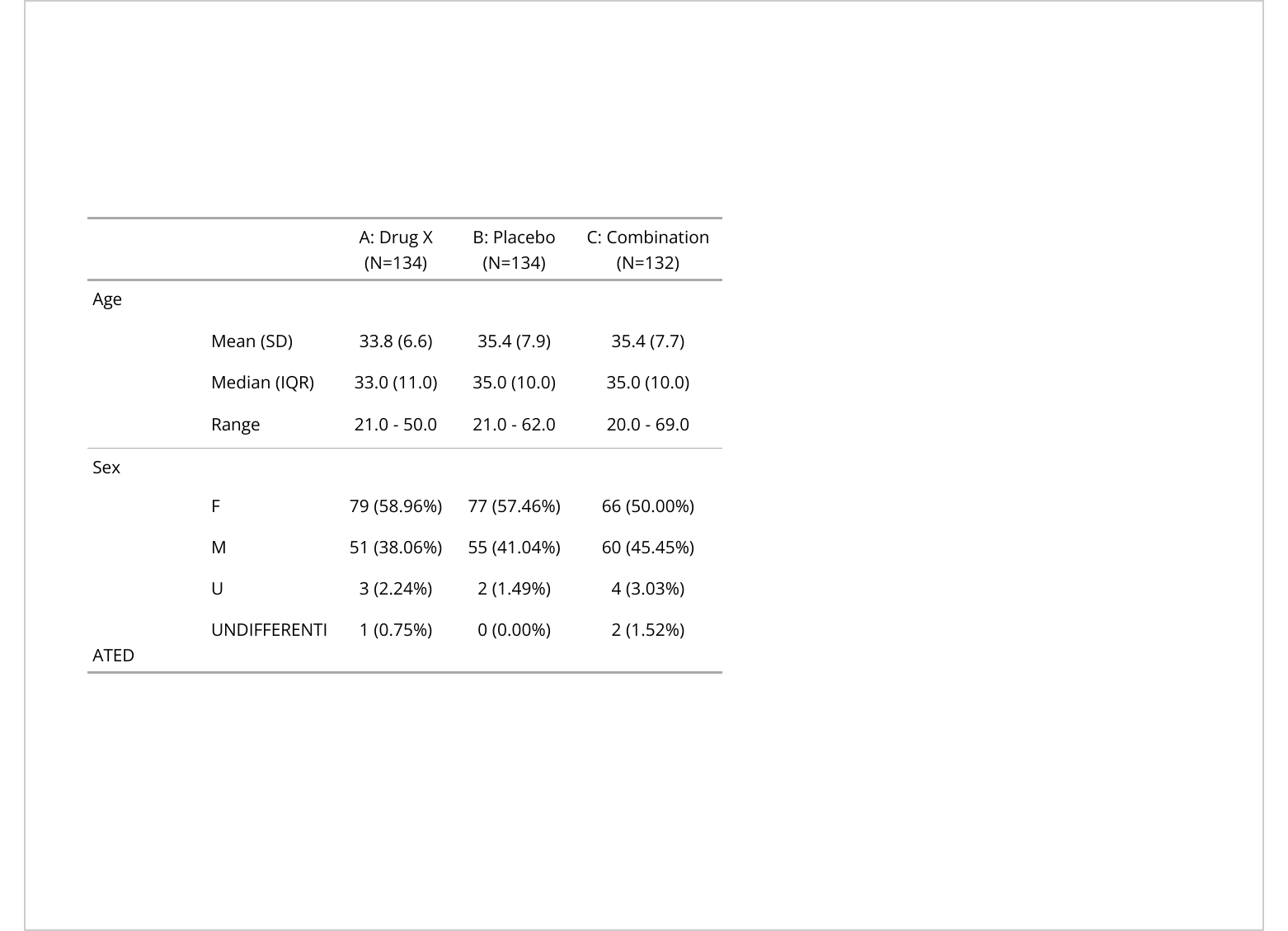
ftA: Drug X | B: Placebo | C: Combination | |
|---|---|---|---|
Age | |||
Mean (SD) | 33.8 (6.6) | 35.4 (7.9) | 35.4 (7.7) |
Median (IQR) | 33.0 (11.0) | 35.0 (10.0) | 35.0 (10.0) |
Range | 21.0 - 50.0 | 21.0 - 62.0 | 20.0 - 69.0 |
Sex | |||
F | 79 (58.96%) | 77 (57.46%) | 66 (50.00%) |
M | 51 (38.06%) | 55 (41.04%) | 60 (45.45%) |
U | 3 (2.24%) | 2 (1.49%) | 4 (3.03%) |
UNDIFFERENTIATED | 1 (0.75%) | 0 (0.00%) | 2 (1.52%) |
Save result in a file
You can quickly save the result:
- in a Microsoft Word with
save_as_docx(), - in a Microsoft PowerPoint with
save_as_pptx(), - in a PNG with
save_as_image()with full support for fonts.
package ‘doconv’ https://cran.r-project.org/package=doconv
R Tables for Regulatory Submissions Working Group
https://rconsortium.github.io/rtrs-wg/
The goal of the working group is to create standards for creating tables that meet the requirements of FDA submission documents, and hence enhance the suitability of R for FDA submissions. […]
Participation in the working group led to several advances in clinical reporting with ‘flextable’.
Two types of table packages
Focused on formatting, with content definition populated from a data.frame (‘flextable’, ‘gt’). The data.frame based approach of ‘flextable’ requires to first build a data.frame that contains all data and then to format it.
data.frame based approach
It sometimes results in code that is too long and painful to write in order to produce the tables in the book.
package | ASCII | HTML | DOCX | RTF | PPTX | GRID | |
|---|---|---|---|---|---|---|---|
flextable | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Focused on functional expression and content definition, with little visual formatting capability (‘tables’, ‘rtables’, ‘tfrmt’, ‘tidytlg’). Packages ‘tables’ and ‘rtables’ are expressive enough to allow these tables to be formulated and can be converted to flextable.
as_flextable approach
Use ‘tables’(Murdoch 2023, website) and get a flextable with as_flextable() function.
Use ‘rtables’(Becker and Waddell 2023, website) and get a flextable with tt_to_flextable() function.
package | ASCII | HTML | DOCX | RTF | PPTX | GRID | |
|---|---|---|---|---|---|---|---|
rtables | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
tables | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Disposition table
This table taken from the book shows the data.frame approach with flextable.
Disposition table preparation
Function tabulator() is used to prepare/structure the table.
The very first step is to create the data.frame that will be used by function tabulator() (40 lines of code with tidyverse).
TRT01A | EOSSTT | DCSREAS | n | percent |
|---|---|---|---|---|
factor | factor | factor | integer | numeric |
A: Drug X | COMPLETED | 68 | 0.5 | |
B: Placebo | COMPLETED | 66 | 0.5 | |
C: Combination | COMPLETED | 73 | 0.6 | |
A: Drug X | ONGOING | 24 | 0.2 | |
B: Placebo | ONGOING | 28 | 0.2 | |
C: Combination | ONGOING | 21 | 0.2 | |
A: Drug X | DISCONTINUED | ADVERSE EVENT | 3 | 0.0 |
A: Drug X | DISCONTINUED | DEATH | 25 | 0.2 |
A: Drug X | DISCONTINUED | LACK OF EFFICACY | 2 | 0.0 |
A: Drug X | DISCONTINUED | PHYSICIAN DECISION | 2 | 0.0 |
n: 47 | ||||
Disposition table creation
ft <- as_flextable(tab,
spread_first_col = TRUE,
columns_alignment = "center"
)
# add '(N=xx)'
TRT_COUNTS <- setNames(
part_header$n_part,
part_header$TRT01A
)
for (TRT_COD in names(TRT_COUNTS)) {
ft <- append_chunks(
x = ft, part = "header", i = 1,
j = tabulator_colnames(
tab,
columns = "content_cell",
TRT01A %in% !!TRT_COD
),
as_chunk(TRT_COUNTS[TRT_COD],
formatter = function(n) sprintf("\n(N=%.0f)", n)
)
)
}
ft <- labelizor(ft,
j = "DCSREAS",
part = "all", labels = function(x) tools::toTitleCase(tolower(x))
) |>
labelizor(
labels = c(Dcsreas = ""),
j = "DCSREAS", part = "header"
) |>
align(
i = ~ !is.na(EOSSTT) | seq_along(EOSSTT) == 1,
j = 1, align = "left"
) |>
prepend_chunks(
i = ~ is.na(EOSSTT),
j = "DCSREAS", as_chunk("\t")
) |>
autofit()A: Drug X | B: Placebo | C: Combination | ||||
|---|---|---|---|---|---|---|
Completed | 68 (50.7%) | 66 (49.3%) | 73 (55.3%) | |||
Ongoing | 24 (17.9%) | 28 (20.9%) | 21 (15.9%) | |||
Discontinued | ||||||
Adverse Event | 3 (2.2%) | 6 (4.5%) | 5 (3.8%) | |||
Death | 25 (18.7%) | 23 (17.2%) | 22 (16.7%) | |||
Adverse Event | 9 (6.7%) | 7 (5.2%) | 10 (7.6%) | |||
Disease Progression | 8 (6.0%) | 6 (4.5%) | 6 (4.5%) | |||
Lost to Follow Up | 2 (1.5%) | 2 (1.5%) | 2 (1.5%) | |||
Missing | 2 (1.5%) | 3 (2.2%) | 2 (1.5%) | |||
Post-Study Reporting of Death | 1 (0.7%) | 2 (1.5%) | 1 (0.8%) | |||
Suicide | 2 (1.5%) | 2 (1.5%) | 1 (0.8%) | |||
Unknown | 1 (0.7%) | 1 (0.7%) | ||||
Lack of Efficacy | 2 (1.5%) | 2 (1.5%) | 3 (2.3%) | |||
Physician Decision | 2 (1.5%) | 3 (2.2%) | 2 (1.5%) | |||
Protocol Violation | 5 (3.7%) | 3 (2.2%) | 4 (3.0%) | |||
Withdrawal by Parent/Guardian | 4 (3.0%) | 2 (1.5%) | 1 (0.8%) | |||
Withdrawal by Subject | 1 (0.7%) | 1 (0.7%) | 1 (0.8%) | |||
Adverse Event Table
This table is using package tables::tabular() to produce a tabular object that will then be transformed into a flextable with function as_flextable().
Adverse Event Table preparation
link: https://rconsortium.github.io/rtrs-wg/commontables.html#tables-5
heading <- tabular(
Heading("")*1*
Heading("")*count ~
Heading()*ARM, data = ex_adsl)
body <- tabular(
Heading("Patients with at least one event")*1*
Heading("")*countpercentid*Arguments(ARM = ARM)*
Heading()*USUBJID +
Heading("Total number of events")*1*Heading("")*1 +
Heading()*AEBODSYS*
(Heading("Patients with at least one event")*
Percent(denom = ARM, fn = countpercentid)*
Heading()*USUBJID +
Heading("Total number of events")*1 +
Heading()*AEDECOD*DropEmpty(which = "row")*
Heading()*Percent(denom = ARM, fn = countpercentid)*
Heading()*USUBJID) ~
Heading()*ARM,
data = ex_adae)
tab <- rbind(heading, body)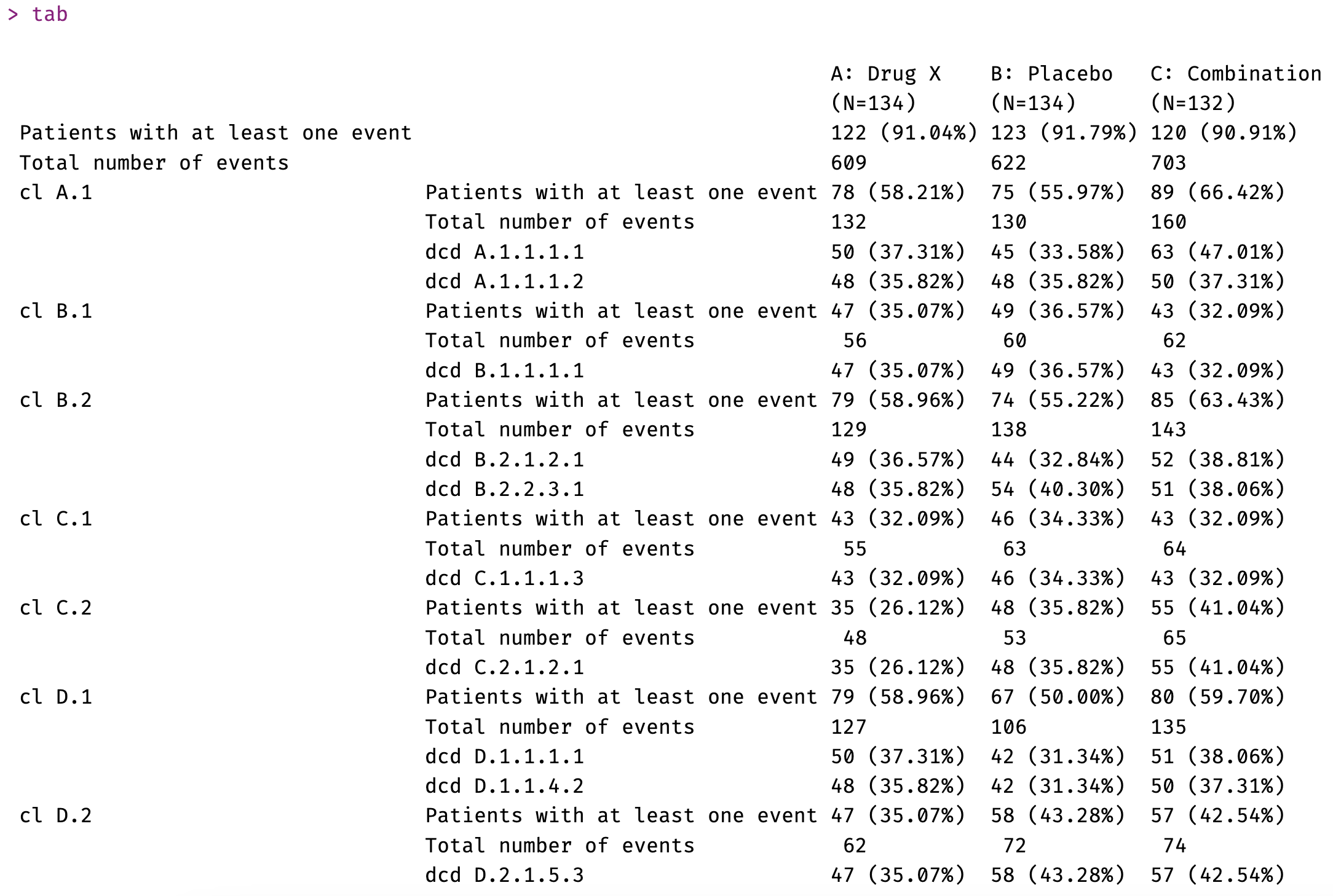
Adverse Event Table creation
‘flextable’ code: https://rconsortium.github.io/rtrs-wg/commontables.html#flextable-5
A: Drug X | B: Placebo | C: Combination | |
|---|---|---|---|
(N=134) | (N=134) | (N=132) | |
Patients with at least one event | 122 (91.04%) | 123 (91.79%) | 120 (90.91%) |
Total number of events | 609 | 622 | 703 |
cl A.1 | |||
Patients with at least one event | 78 (58.21%) | 75 (55.97%) | 89 (66.42%) |
Total number of events | 132 | 130 | 160 |
dcd A.1.1.1.1 | 50 (37.31%) | 45 (33.58%) | 63 (47.01%) |
dcd A.1.1.1.2 | 48 (35.82%) | 48 (35.82%) | 50 (37.31%) |
cl B.1 | |||
Patients with at least one event | 47 (35.07%) | 49 (36.57%) | 43 (32.09%) |
Total number of events | 56 | 60 | 62 |
dcd B.1.1.1.1 | 47 (35.07%) | 49 (36.57%) | 43 (32.09%) |
cl B.2 | |||
Patients with at least one event | 79 (58.96%) | 74 (55.22%) | 85 (63.43%) |
Total number of events | 129 | 138 | 143 |
dcd B.2.1.2.1 | 49 (36.57%) | 44 (32.84%) | 52 (38.81%) |
dcd B.2.2.3.1 | 48 (35.82%) | 54 (40.30%) | 51 (38.06%) |
cl C.1 | |||
Patients with at least one event | 43 (32.09%) | 46 (34.33%) | 43 (32.09%) |
Total number of events | 55 | 63 | 64 |
dcd C.1.1.1.3 | 43 (32.09%) | 46 (34.33%) | 43 (32.09%) |
cl C.2 | |||
Patients with at least one event | 35 (26.12%) | 48 (35.82%) | 55 (41.04%) |
Total number of events | 48 | 53 | 65 |
dcd C.2.1.2.1 | 35 (26.12%) | 48 (35.82%) | 55 (41.04%) |
cl D.1 | |||
Patients with at least one event | 79 (58.96%) | 67 (50.00%) | 80 (59.70%) |
Total number of events | 127 | 106 | 135 |
dcd D.1.1.1.1 | 50 (37.31%) | 42 (31.34%) | 51 (38.06%) |
dcd D.1.1.4.2 | 48 (35.82%) | 42 (31.34%) | 50 (37.31%) |
cl D.2 | |||
Patients with at least one event | 47 (35.07%) | 58 (43.28%) | 57 (42.54%) |
Total number of events | 62 | 72 | 74 |
dcd D.2.1.5.3 | 47 (35.07%) | 58 (43.28%) | 57 (42.54%) |
Concomitant Medications Table
This table is using package ‘rtables’ to produce an object that will then be transformed into a flextable with function rtables::tt_to_flextable().
Concomitant Medications preparation
link: https://rconsortium.github.io/rtrs-wg/commontables.html#rtables-7
lyt <- basic_table(
show_colcounts = TRUE) |>
split_cols_by("TRT01A") |>
analyze("EOSSTT", top_afun) |>
split_rows_by("EOSSTT",
split_fun =
keep_split_levels("DISCONTINUED")
) |>
analyze("DCSREAS", count_pct_afun) |>
split_rows_by("DCSREAS",
split_fun =
keep_split_levels("DEATH")
) |>
analyze("DTHCAUS", count_pct_afun)
tab <- build_table(lyt, adsl) A: Drug X B: Placebo C: Combination
(N=134) (N=134) (N=132)
——————————————————————————————————————————————————————————————————————————
Completed 68 (50.7%) 66 (49.3%) 73 (55.3%)
Ongoing 24 (17.9%) 28 (20.9%) 21 (15.9%)
DISCONTINUED
ADVERSE EVENT 3 (2.2%) 6 (4.5%) 5 (3.8%)
DEATH 25 (18.7%) 23 (17.2%) 22 (16.7%)
LACK OF EFFICACY 2 (1.5%) 2 (1.5%) 3 (2.3%)
PHYSICIAN DECISION 2 (1.5%) 3 (2.2%) 2 (1.5%)
PROTOCOL VIOLATION 5 (3.7%) 3 (2.2%) 4 (3.0%)
WITHDRAWAL BY PARENT/GUARDIAN 4 (3.0%) 2 (1.5%) 1 (0.8%)
WITHDRAWAL BY SUBJECT 1 (0.7%) 1 (0.7%) 1 (0.8%)
DEATH
ADVERSE EVENT 9 (6.7%) 7 (5.2%) 10 (7.6%)
DISEASE PROGRESSION 8 (6.0%) 6 (4.5%) 6 (4.5%)
LOST TO FOLLOW UP 2 (1.5%) 2 (1.5%) 2 (1.5%)
MISSING 2 (1.5%) 3 (2.2%) 2 (1.5%)
Post-study reporting of death 1 (0.7%) 2 (1.5%) 1 (0.8%)
SUICIDE 2 (1.5%) 2 (1.5%) 1 (0.8%)
UNKNOWN 1 (0.7%) 1 (0.7%) 0 (0.0%) Concomitant Medications creation
Fonts, table layout and few other details need to be changed to produce a nice flextable.
A: Drug X | B: Placebo | C: Combination | |
|---|---|---|---|
(N=134) | (N=134) | (N=132) | |
Completed | 68 (50.7%) | 66 (49.3%) | 73 (55.3%) |
Ongoing | 24 (17.9%) | 28 (20.9%) | 21 (15.9%) |
DISCONTINUED | |||
ADVERSE EVENT | 3 (2.2%) | 6 (4.5%) | 5 (3.8%) |
DEATH | 25 (18.7%) | 23 (17.2%) | 22 (16.7%) |
LACK OF EFFICACY | 2 (1.5%) | 2 (1.5%) | 3 (2.3%) |
PHYSICIAN DECISION | 2 (1.5%) | 3 (2.2%) | 2 (1.5%) |
PROTOCOL VIOLATION | 5 (3.7%) | 3 (2.2%) | 4 (3.0%) |
WITHDRAWAL BY PARENT/GUARDIAN | 4 (3.0%) | 2 (1.5%) | 1 (0.8%) |
WITHDRAWAL BY SUBJECT | 1 (0.7%) | 1 (0.7%) | 1 (0.8%) |
DEATH | |||
ADVERSE EVENT | 9 (6.7%) | 7 (5.2%) | 10 (7.6%) |
DISEASE PROGRESSION | 8 (6.0%) | 6 (4.5%) | 6 (4.5%) |
LOST TO FOLLOW UP | 2 (1.5%) | 2 (1.5%) | 2 (1.5%) |
MISSING | 2 (1.5%) | 3 (2.2%) | 2 (1.5%) |
Post-study reporting of death | 1 (0.7%) | 2 (1.5%) | 1 (0.8%) |
SUICIDE | 2 (1.5%) | 2 (1.5%) | 1 (0.8%) |
UNKNOWN | 1 (0.7%) | 1 (0.7%) | 0 (0.0%) |
Pagination
The pagination of tables allows you to control their position in relation to page breaks. Packages ‘tables’, ‘rtables’ and ‘flextable’ implement their pagination vision.
Pagination with ‘flextable’
When working with Word or RTF, it is possible to prevents breaks between tables rows you want to stay together. Function paginate() let you define this pagination.
From https://ardata-fr.github.io/flextable-book/layout.html#pagination

Pagination with ‘rtables’
tbl_list <- basic_table() |>
split_cols_by("ARM") |>
split_rows_by(
"STRATA1",
split_fun = keep_split_levels(c("A", "B")),
page_by = TRUE, page_prefix = "Stratum") |>
split_rows_by(
"RACE",
split_fun = keep_split_levels(c("ASIAN", "WHITE"))) |>
summarize_row_groups() |>
analyze("AGE", afun = function(x, ...)
in_rows(
"mean (sd)" = rcell(c(mean(x), sd(x)), format = "xx.x (xx.x)"),
"range" = rcell(range(x), format = "xx.x - xx.x")
)) |>
build_table(formatters::ex_adsl) |>
paginate_table(lpp = 20)Now let’s use ‘officer’ to add tables into a Word document, one table per page.

Pagination with ‘tables’
Let’s create a quick example with ‘tables’.
Now let’s use ‘officer’ to add tables into a Word document, one table per page.
Advanced Word features
We’re going to take a look at some of the features that go well with clinical reporting and Word file production:
- adding computed fields
- section management with officer.
Word Computed fields and flextable
doc <- read_docx()
for (spl in splits) {
ft <- as_flextable(tab[spl, ],
spread_first_col = TRUE
) |>
add_footer_lines(
values =
as_paragraph(
"p. ",
as_word_field(x = "Page", width = .05),
" on ", as_word_field(x = "NumPages", width = .05)
)
)
doc <- body_add_flextable(doc, ft) |>
body_add_break()
}
doc <- body_remove(doc) |>
print(
target = "assets/files/tables-fields.docx"
)Use section headers and footers
header_default <-
block_list(
fpar(ftext("p. "),
run_word_field(field = "Page"),
" on ",
run_word_field(field = "NumPages")))
footer_default <-
block_list(
fpar(
external_img(src = "assets/img/r-in-pharma-logo.png")))
ps <- prop_section(
header_default = header_default,
footer_default = footer_default)
doc <- read_docx()
for(spl in splits) {
ft <- as_flextable(tab[spl,],
spread_first_col = TRUE)
doc <- body_add_flextable(doc, ft)|>
body_add_break()
}
doc <- body_remove(doc) |>
body_set_default_section(value = ps) |>
print(target = "assets/files/tables-sections.docx")Thank you
- Tables in Clinical Trials with R: https://rconsortium.github.io/rtrs-wg/
- The flextable book: https://ardata-fr.github.io/flextable-book/
- The flextable gallery: https://ardata.fr/flextable-gallery/
- source of the presentation: https://github.com/ardata-fr/r-in-pharma-2023-10-24
- presentation: https://ardata.fr/r-in-pharma-2023-10-24
References: